About the site
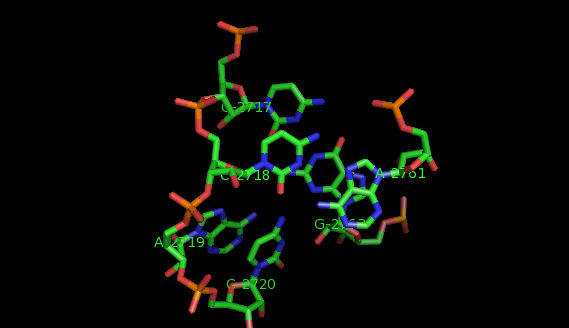
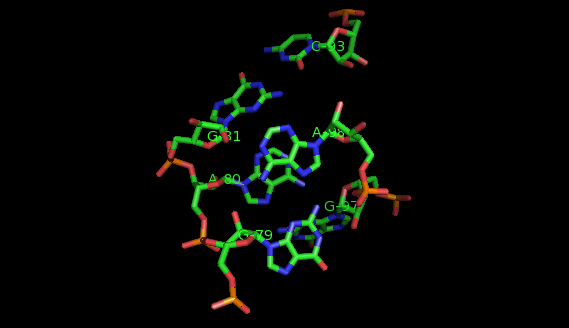
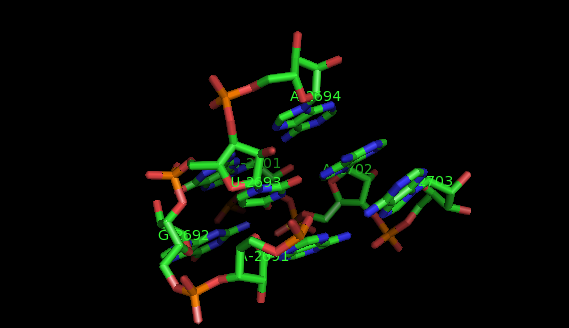
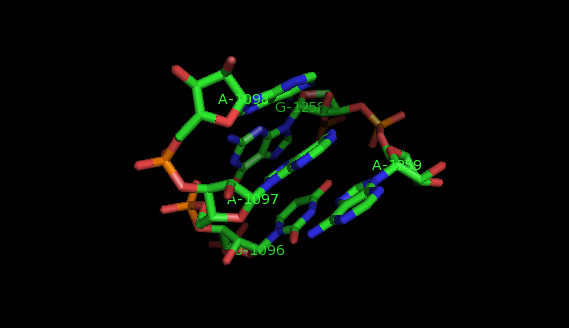
This website offers a package of programs aimed to extract recurrent tertiary motifs in RNA structures (introductory information on tertiary motifs is provided by the What are motifs? link on the left panel).The motifs considered are of two types: local motifs and interation motifs. Local motifs are embedded within secondary structure elements (internal, junction, terminal loops) while interaction motifs link two or more secondary structure elements and/or helices.
The method implemented is based on a graph representation of the RNA tertiary structure. Practically, the RNA structure (PDB file) is analysed by the annotation program FR3D. The result of this analysis is a set of lists of bases' interactions (LW base pairings, stacking interactions, backbone relations, etc.). These lists of interactions provide a graph model where the vertices represent nucleotides labeled by their sequence letter and residue number, and the edges represent base pairs labeled by their type of interaction. In our model, a recurrent tertiary motif is a subgraph of the graph of the tertiary structure which occurs more than once in the structure. These occurrences are topologically similar and not necessarily identical.
According to the type of motif you are interested in (local or interaction motif), visit the corresponding tab (LOCAL MOTIFS/INTERACTION MOTIFS) to see how to use my programs.
When using the website, authors should cite the following publication:
Automated motif extraction and classification in RNA tertiary structures
RNA. 2008 Dec;14(12):2489-97
The images of the motifs featured in this site were produced with Pymol using public PDB files downloaded from the Protein Data Bank.
|
|
|

|