LOCAL MOTIFS
In the following, we assume that you have downloaded the Rna3Dmotif binaries archive and uncompressed it according to the instructions given in the DOWNLOAD section.Cataloguing
Local motifs are embedded within secondary structure elements (internal, terminal, junction loops). So the first step consists of identifying these elements in the graph model of the RNA structure. The result of this step is called Catalogue.
For example, suppose that you want to build the Catalogues of two structures:
- 5S rRNA of H. marismortui (PDB: 1S72, Chain: 9),
- 23S rRNA of H. marismortui (PDB: 1S72, Chain: 0).
After executing the Catalog program, you get this result (see the User's Help for the notations used).
Identifying recurrent local motifs
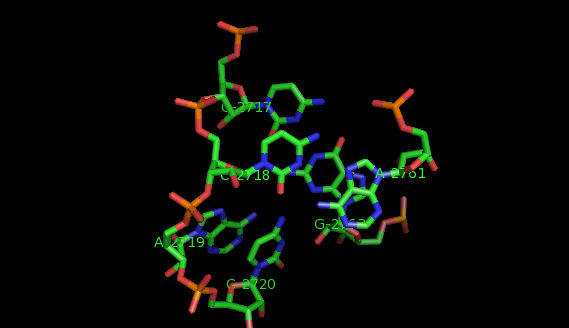
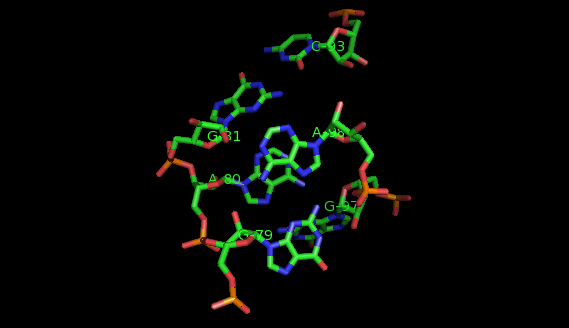
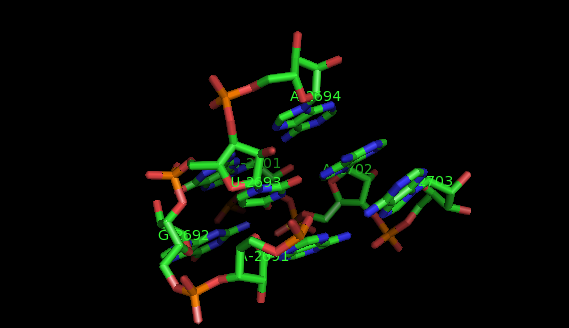
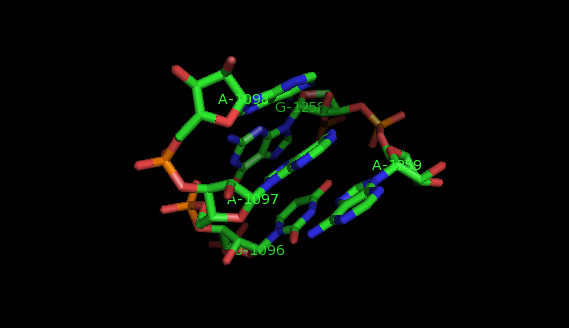
Suppose now that you want to identify recurrent local motifs present in two RNA chains, say 5S and 23S rRNAs of H. marismortui (PDB: 1S72, Chains: 9 and 0). After executing the Simlocmot program, you get this spreadsheet file. You sort this file by the last column (i.e. the non-canonic labels of the LECNS) to get the clusters of potential recurrent motifs. For example, in our sorted file, these lines give the occurrences of a potential recurrent motif with the non-canonical labels "eilm". These occurrences must be superposed in 3D to make sure that they are biologically valid. In our example, all the occurrences were successfully superposed. The motif in question is the Sarcin-ricin motif (6 occurrences in 23S rRNA and 1 occurrence in 5S rRNA).
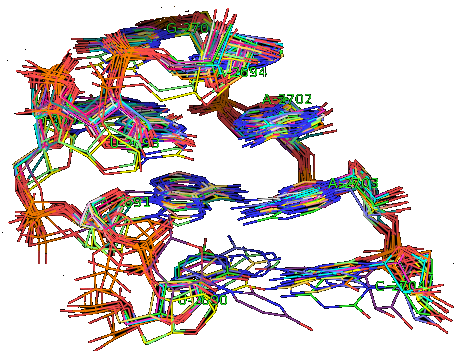
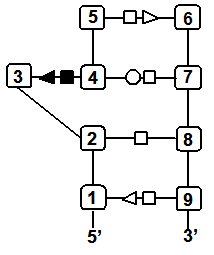
Finally, you may wish to compare two specific secondary structure elements. In this case, you invoke the Lecns program which inputs two identifiers of secondary structure elements and outputs, if it exists, their largest extensible common non-canonical substructure (LECNS) in the form of a mapping of the bases and basepairs. This program is useful to detect similarities between two secondary structure elements of a particular interest for the user. For example, this mapping displays the structural similarities between a secondary structure element containing a sarcin motif in 5S rRNA of H. marismortui and a secondary structure element containing an E-loop motif in 16S rRNA of T. thermophilus.
|
|
|

|